COLONY COUNT
|
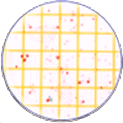
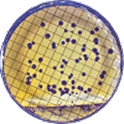
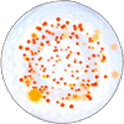
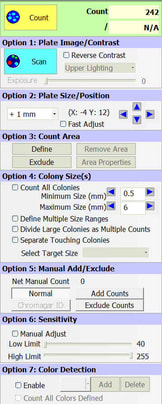
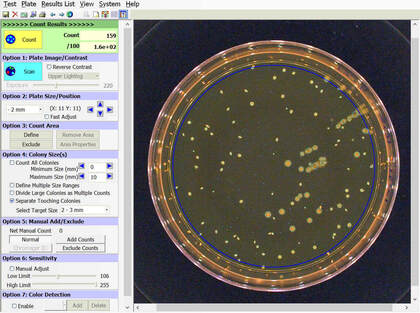
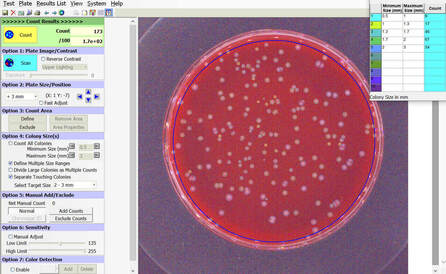
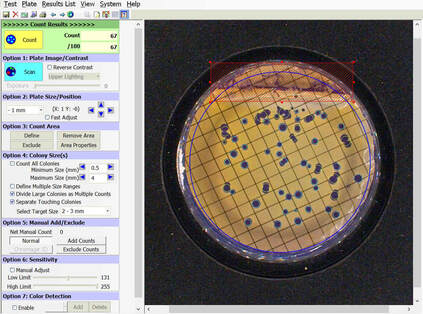
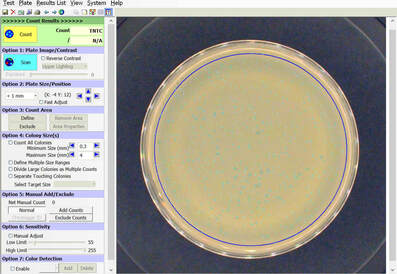
BIOMIC V3 automatically counts bacterial, yeast, and mould colonies and virus plaque assays on agar plates with color digital imaging technology. This saves 99% of reading time and eliminates subjective variation and transcription errors. Plate images are displayed in high resolution with counting tools and options available on-screen. Parameters can be defined to optimize counting performance and accuracy. Plate images and results are saved for future review. Chromogenic agar ID is included with the Colony Count module. Learn more > |
Colony Count - Plate Types
Colony Count - Reading Options
|
Select the video above to view (2:40)
Select the icon on the bottom right of the video for full screen mode.
Click here to view all videos >
Select the icon on the bottom right of the video for full screen mode.
Click here to view all videos >
Purpose
The purpose of this software module is to count colony forming units (cfu) of bacteria or yeast in a test sample or aliquot, that is a portion of a larger quantity of liquid, material or whole. From this sample, one should be able to estimate the relative quantity of bacteria or yeast in the whole, by multiplying the sample count (cfu) by the dilution factor. This count must be considered an approximation for many reasons, including limitations of the: 1) sampling method, 2) number of organisms in each sample, 3) contamination, transportation, storage, 4) assay methodology, 5) software threshold settings for colony size and contrast, and 6) human eye/camera resolution. For these reasons, an enlarged image of each test plate is provided on screen for a microbiologist to review, and edit as required.
Limitations
The purpose of this software module is to count colony forming units (cfu) of bacteria or yeast in a test sample or aliquot, that is a portion of a larger quantity of liquid, material or whole. From this sample, one should be able to estimate the relative quantity of bacteria or yeast in the whole, by multiplying the sample count (cfu) by the dilution factor. This count must be considered an approximation for many reasons, including limitations of the: 1) sampling method, 2) number of organisms in each sample, 3) contamination, transportation, storage, 4) assay methodology, 5) software threshold settings for colony size and contrast, and 6) human eye/camera resolution. For these reasons, an enlarged image of each test plate is provided on screen for a microbiologist to review, and edit as required.
Limitations
- Sampling Method: If numerous samples/aliquots taken from the same larger whole vary significantly in number of colonies counted, it may mean organisms are not evenly distributed in the whole. If higher accuracy is desired, more test samples may be taken from the whole, perhaps from different areas in the whole. Debris in samples may also render counting difficult.
- Number of Organisms in Each Test Sample: If the number of organisms in a test sample is too numerous to count (TNTC), it may be accurate to report it as such. Optimal counting accuracy generally occurs between one to 99 colonies per plates. At higher #’s, for greater accuracy, the original sample may be diluted 1/5 to 1/10, re-plated, re-incubated, and re-counted. Consider the purpose and limitations of this test count before going to extremes to provide accuracy.
- Contamination, Transportation, Storage is Critical: The microbiologist should verify test samples are always collected with careful technique, or contamination may result. All samples must be transported and stored under appropriate conditions before testing, usually chilled/refrigerated. If not, counts will not reflect the whole. Fungal mold colonies may overgrow a plate and render accurate counts difficult or impossible. Agar surface counts are not designed to count molds; if you anticipate counting mold colonies, an agar overlay method may be considered to result in more defined mold colonies.
- Assay Methodology: There are limitations to the accuracy of this assay method. For example: some bacteria or yeast may tend to grow in clumps; a clump will result in one cfu, just as a single bacterial cell may be one cfu. Each sample should be well spread on agar surface. Even with careful attention to assay technique by a microbiologist, other limitations may be significant.
- Software Threshold Settings for Colony Size and Contrast: If many colonies are on the threshold of size or contrast settings in software, counts will tend to vary more.
- Human Eye/Camera Resolution: Different human eyes have different abilities to discriminate what is a true colony versus debris, relative to the camera. Some discriminating points may be difficult or impossible to program into software logic.